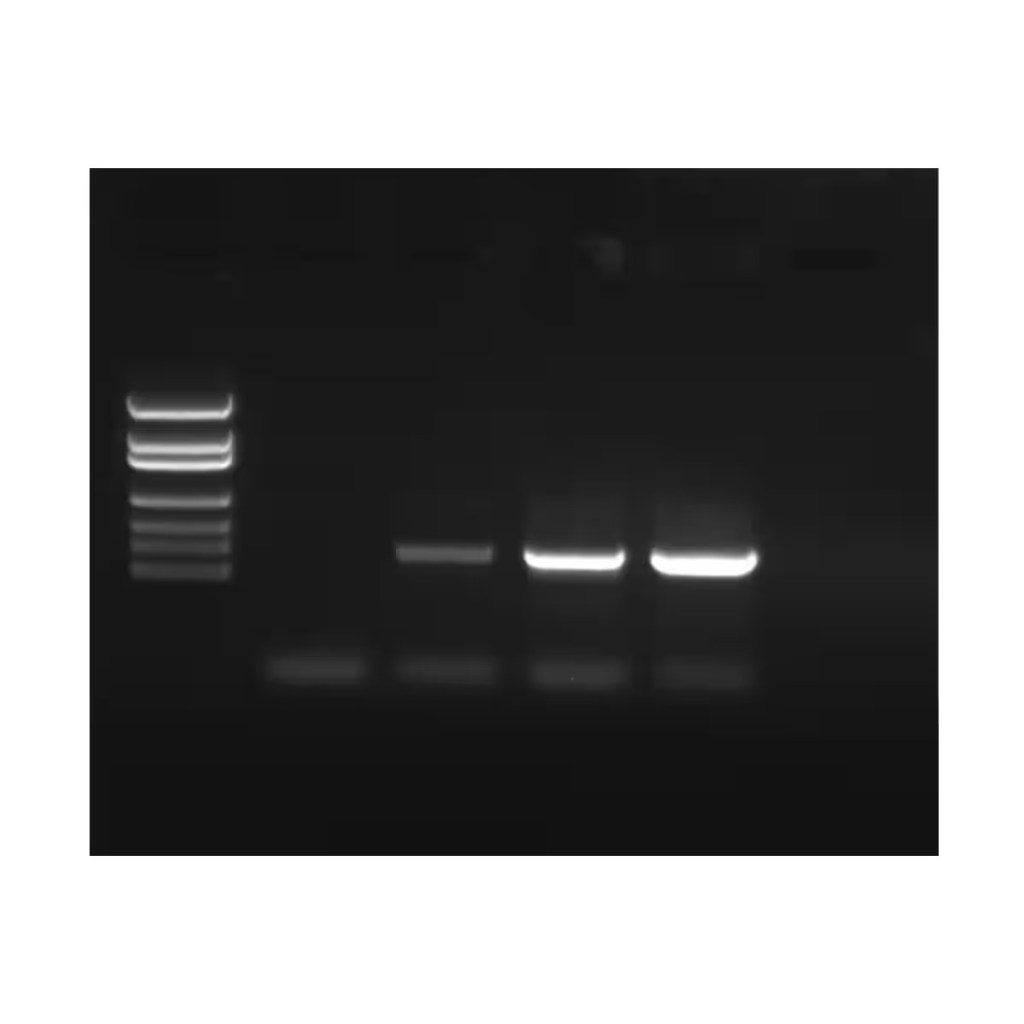
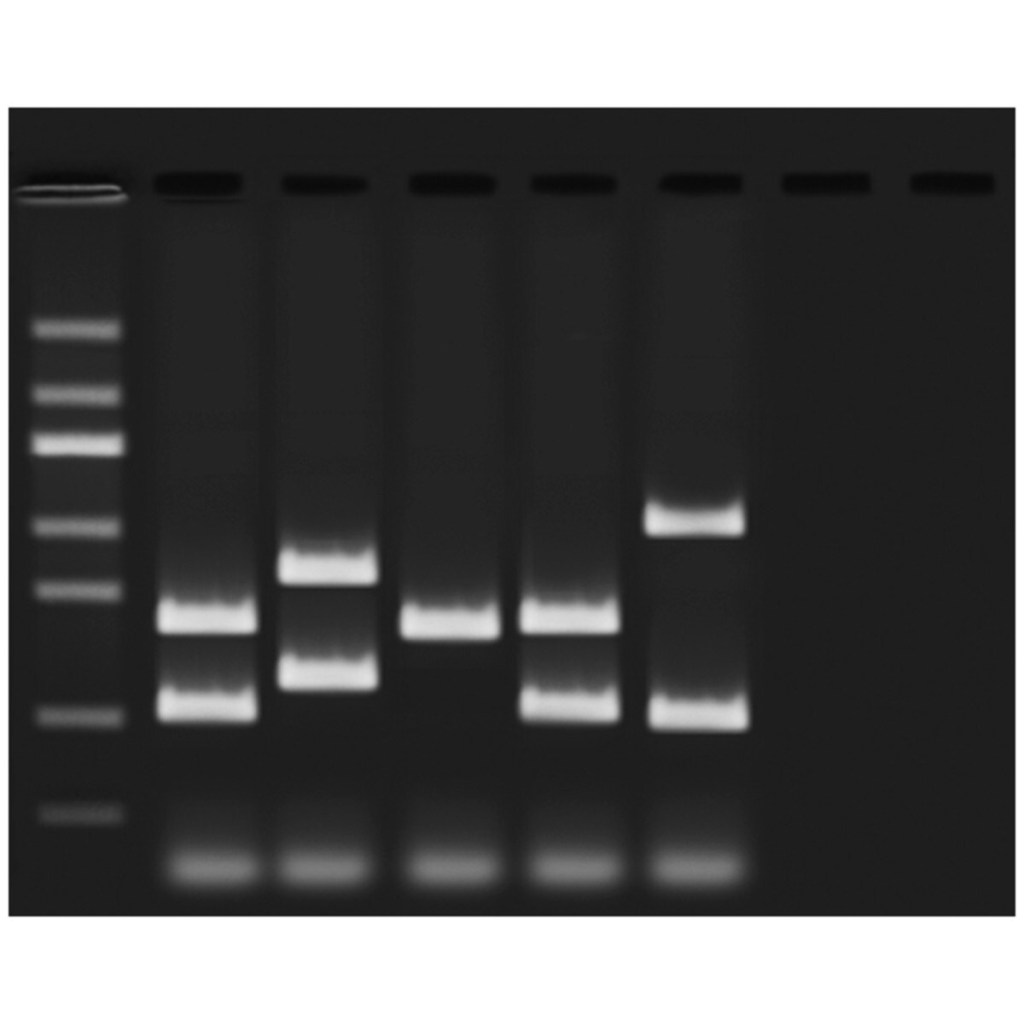
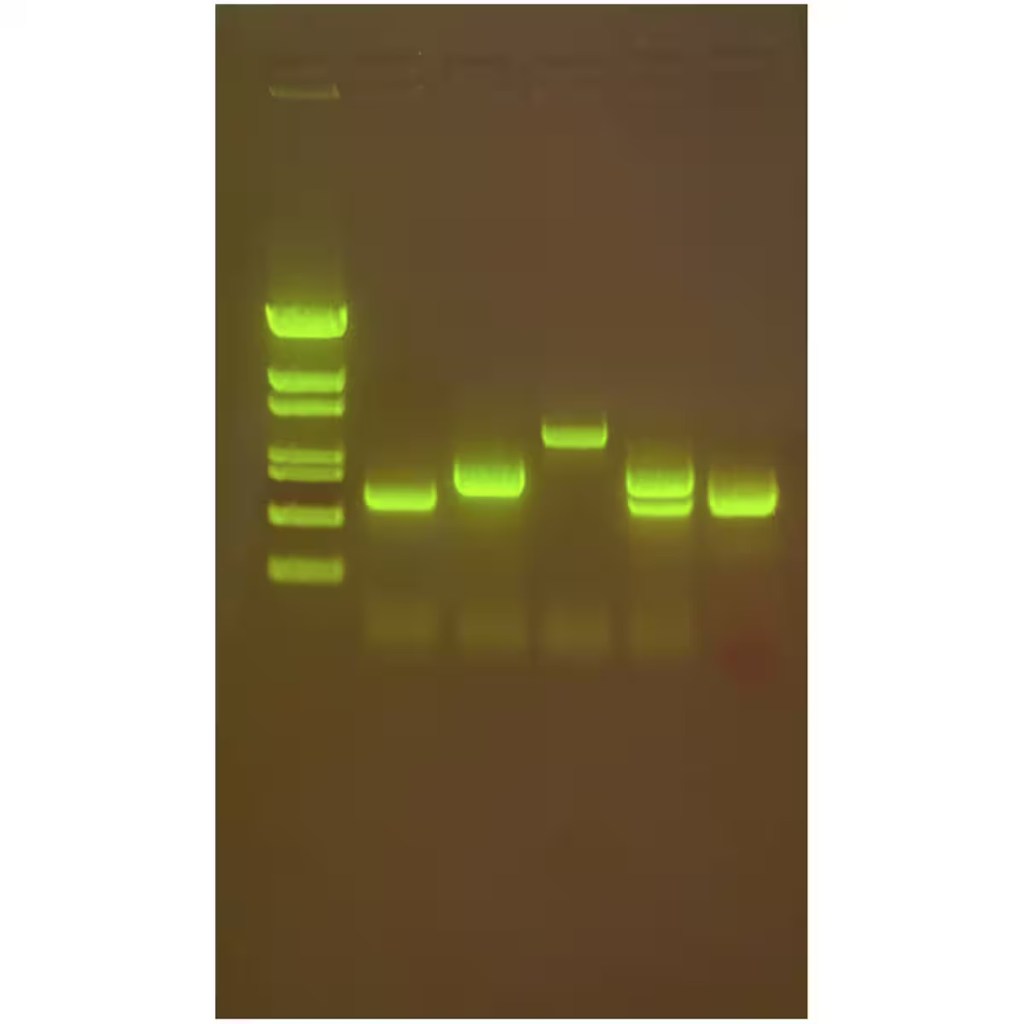
Polymerase Chain Reaction (PCR) is a lab technique used to make millions of copies of a specific piece of DNA. PCR works by heating the DNA to separate the strands, then cooling it so short pieces of DNA called primers can attach to the target sequence. A special enzyme called DNA polymerase then builds new strands of DNA. This cycle is repeated many times, doubling the amount of DNA each time. Today, Edvotek brings you a guide to everything related to PCR primers!
PCR primers are short pieces of DNA that help start the copying process in PCR. They are designed to be complimentary to the beginning and end of the DNA section scientists want to copy. This means when the template DNA is separated into single strands, each primer attaches to each strand of DNA to allow for the polymerase to replicate the area between the two primers. Without primers, the polymerase wouldn’t have a place to start, so they are a key part of making PCR work. Here are some of the major criteria for designing PCR primers:
- Primer Length: Primers should be 18-25 nucleotides long. They should be long enough to ensure specific binding to the targeted region but short enough to bind efficiently.
- Melting Temperature (Tm): The melting temperature is the temperature at which half the primer strands will separate into single stranded pieces of DNA. This property is dependent on the primer’s nucleotide sequence. To ensure your primers bind to the template strands with the same affinity, they should have Tm values within 5 ˚C of each other. Furthermore, you should generally design primers to have a Tm between 55 – 65 ˚C. New England Biolabs has a great online Tm calculator for checking this.
- GC content: Considering the amount of cytosine (C) and guanine (G) nucleotides in your primer is also very important. C and G form three hydrogen bonds between them when paired as complimentary bases in double stranded DNA. C and G pairs require more energy to break apart than A and T pairs which only contain two hydrogen bonds. Because GC pairs have a greater effect on primer binding strength they should ideally make up 40 – 60% of the nucleotides in your primer – not too high and not too low. Additionally, having 1-2 GC pairs at the 3’ end of your primers will improve binding.
- Site specificity: As previously mentioned, your primers should be designed to bind to specific sites that flank the desired region to be amplified. There are several tools online that can help you with this such as The National Institute of Health’s primer design tool. Online primer design tools are very useful for ensuring that your primers aren’t targeting any other regions on the template you don’t want amplified. This program can also predict secondary structures within the template DNA such as hairpins which could potentially prevent primer binding or result in non-specific products.
- Primer pairs should not have complimentary regions: If your primer pairs have nucleotide stretches that are complementary to each other, they will form a complex with each other known as a primer dimer. Prevent complementary regions in your primers to ensure they only target the template DNA.
We hope you have found this information useful and keep it in mind for future primer designs! If you want to learn more about PCR we have more information on our website and on our youtube channel. Interested in getting hands-on experience with PCR? Check out some of our kits below: